Abstract
Background: Waldenström's Macroglobulinemia (WM) is a distinct, indolent, IgM lymphoplasmacytic lymphoma with a heterogeneous clinical course. Genomic characterization of WM has revealed recurrent somatic mutations in MYD88 (>95% patients) and CXCR4 (>30% patients) genes, and deletions involving chromosome 6q (del6q, ~50% patients), among other alterations. MYD88 L265P is considered to be the tumor-initiating event that provides an advantage for B-cell clonal selection and predisposes the malignant clone to further genetic alterations, leading to full-blown lymphoma development. However, the highly variable disease evolution cannot be explained solely by the pattern of genomic alterations. The cell of origin, the distribution in individual tumor cells and clones and how these interact may influence the oncogenic process. Here, we have performed a single cell study of WM patients to characterize the complete tumor architecture, determine the timing of the alterations and provide information about mutations that cooperate in the development of the disease.
Methods: Bone marrow mononuclear cells from eight WM patients (five at diagnosis and three at progression) were preserved in fetal bovine serum at -80º until their use. We designed a Tapestri Single-Cell DNA Custom Panel (Mission Bio) with the hotspot regions of 20 genes (including those for the assessment of 6q deletion), and an antibody-oligo conjugated panel to identify the different cell populations of WM by targeting the following surface proteins: CD19, CD20, CD34, CD38, and CD138. Libraries were sequenced on a NextSeq 1000/2000 sequencer (Illumina) using 2x150 bp cycles. Sequencing data were processed using Tapestri Pipeline v2, which includes: (1) adapter-trimming using Cutadapt, (2) reference genome alignment to hg19, (3) cellular barcode demultiplexing, and (4) cell-based genotype calling using GATK/Haplotypecaller. Loom and h5 files were analyzed with Tapestri Insights v2.2 (variant selection) and the Python-based Mosaic package (multiomics analysis and data visualization). Only cells with complete genotype information of the variants used for downstream analysis were included.
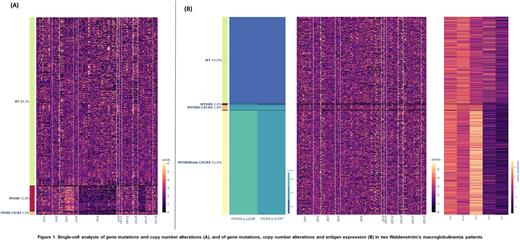
Results: A median number of 5294 cells from the eight patients were analyzed. MYD88 L265P was the most clonal alteration at diagnosis, thus supporting its role as a driver event. Clonal selection could be inferred in progression samples, as the secondary alterations were generally present in the same cells as MYD88 mutation. Del6q was found in 5/8 patients, showing mutual exclusivity with CXCR4 mutations (except in one patient), which suggests there might be at least two pathways that promote disease progression. In the patient in whom both alterations co-existed, there were two clearly different subclones that could be responsible for progression: (1) MYD88-del6q-del17p-gain3q and (2) MYD88-CXCR4-del6q (Figure 1A). Deletions of two negative regulators of the NF-κB signaling pathway, TRAF3 and TNFAIP3 (del6q), also co-occurred in the same clone in one patient. In another patient, homozygous mutated MYD88 was observed due to an event of acquired uniparental disomy, which arose after CXCR4 alteration. This subclone also showed higher expression of CD19 (Figure 1B). Data regarding protein expression will be further analyzed to be presented at the meeting.
Conclusions: Single cell sequencing techniques allow to characterize tumor heterogeneity and elucidate the order of appearance and co-dependency of alterations, which could shed light on the putative mechanisms leading to progression. WM evolution seems to be consistent with a branching model in which only clones containing MYD88 mutation give rise to more aggressive populations by acquiring new aberrations.
Disclosures
García-Sanz:Janssen: Honoraria, Other: Travel support, Research Funding; BeiGene: Honoraria, Other: Travel Support; Gilead: Honoraria, Research Funding; Astellas: Honoraria, Research Funding; Amgen: Honoraria; Takeda: Honoraria, Research Funding; GSK: Honoraria, Other: Travel Support; Astra Zeneca: Honoraria; In Vivo Scribe: Patents & Royalties: Indirect perception, Euroclonality primers; Novartis: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.